|
|
@@ -68,9 +68,10 @@
|
|
|
<h1>rcsb-saguaro-3D</h1>
|
|
|
</a>
|
|
|
<p>RCSB Saguaro Web 3D is an open-source library built on the top of the <a href="https://rcsb.github.io/rcsb-saguaro">RCSB Saguaro 1D Feature Viewer</a>
|
|
|
- and <a href="https://github.com/rcsb/rcsb-molstar">RCSB Molstar</a> designed to display protein features at the <a href="https://www.rcsb.org">RCSB Web Site</a>. The package collects protein annotations from the
|
|
|
- <a href="https://1d-coordinates.rcsb.org">1D Coordinate Server</a> and the main <a href="https://data.rcsb.org">RCSB Data API</a> and generates Protein
|
|
|
- Feature Summaries. The package allows access to RCSB Saguaro and Molstar methods to add or change displayed data. </p>
|
|
|
+ and <a href="https://github.com/rcsb/rcsb-molstar">RCSB Molstar</a> designed to display protein features at the <a href="https://www.rcsb.org">RCSB Web Site</a>.
|
|
|
+ The package collects protein annotations from the <a href="https://1d-coordinates.rcsb.org">1D Coordinate Server</a>
|
|
|
+ and the main <a href="https://data.rcsb.org">RCSB Data API</a> and generates Protein Feature Summaries.
|
|
|
+ The package allows access to RCSB Saguaro and Molstar methods to add or change the displayed data. </p>
|
|
|
<div id="pfv"></div>
|
|
|
<script crossorigin src="https://unpkg.com/react@17/umd/react.production.min.js"></script>
|
|
|
<script crossorigin src="https://unpkg.com/react-dom@17/umd/react-dom.production.min.js"></script>
|
|
|
@@ -368,9 +369,18 @@ document.addEventListener("DOMContentLoaded", function (event) {
|
|
|
<a href="#main-classes-and-methods" id="main-classes-and-methods" style="color: inherit; text-decoration: none;">
|
|
|
<h3>Main Classes and Methods</h3>
|
|
|
</a>
|
|
|
- <p>Class <strong><code>RcsbFv3DAssembly</code></strong> file <code>src/RcsbFv3D/RcsbFv3DAssembly.tsx</code> builds a predefined view for PDB entries. This is the methods used in the RCSB PDB web portal
|
|
|
- (ex: <a href="https://www.rcsb.org/3d-sequence/4HHB">4hhb</a>). Source code example can be found in <code>src/examples/assembly/index.ts</code>.</p>
|
|
|
+ <p>Class <strong><code>RcsbFv3DAssembly</code></strong> (<code>src/RcsbFv3D/RcsbFv3DAssembly.tsx</code>) builds a predefined view for PDB entries. This method is used in the RCSB PDB web portal
|
|
|
+ to display 1D features on PDB entries (ex: <a href="https://www.rcsb.org/3d-sequence/4HHB">4hhb</a>). Source code example can be found in <code>src/examples/assembly/index.ts</code>.</p>
|
|
|
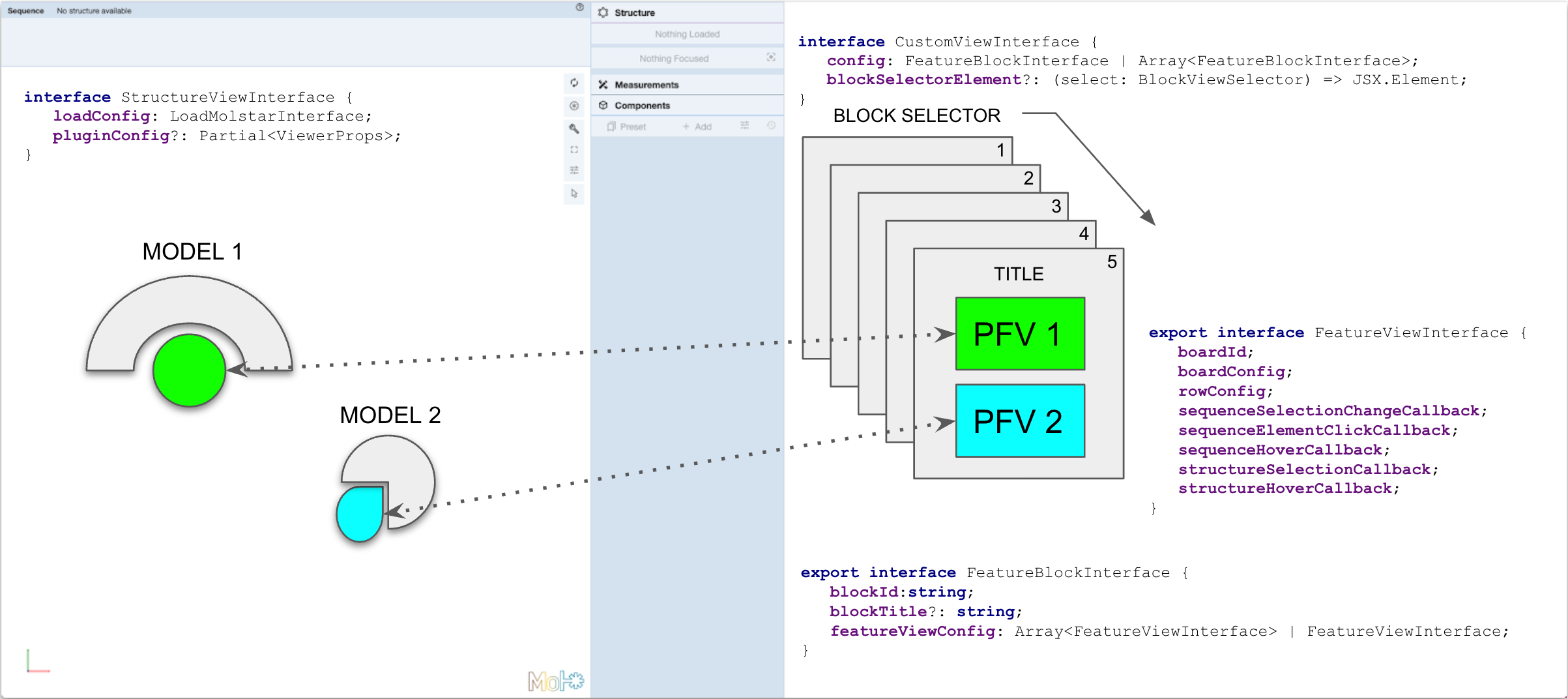
<p>Class <strong><code>RcsbFv3DCustom</code></strong> file <code>src/RcsbFv3D/RcsbFv3DCustom.tsx</code> builds a customized view between one or more feature viewers and a single Molstar plugin.</p>
|
|
|
+ <pre><code class="language-typescript"><span class="hljs-keyword">interface</span> RcsbFv3DCustomInterface <span class="hljs-keyword">extends</span> RcsbFv3DAbstractInterface {
|
|
|
+ <span class="hljs-attr">structurePanelConfig</span>: RcsbFvStructureInterface;
|
|
|
+ sequencePanelConfig: {
|
|
|
+ <span class="hljs-attr">config</span>: CustomViewInterface;
|
|
|
+ title?: <span class="hljs-built_in">string</span>;
|
|
|
+ subtitle?: <span class="hljs-built_in">string</span>;
|
|
|
+ };
|
|
|
+}</code></pre>
|
|
|
+ <p><img src=".github/img/config_img.png?raw=true" alt="Alt text" title="Custom config schema"></p>
|
|
|
<a href="#contributing" id="contributing" style="color: inherit; text-decoration: none;">
|
|
|
<h2>Contributing</h2>
|
|
|
</a>
|
|
|
@@ -631,4 +641,4 @@ document.addEventListener("DOMContentLoaded", function (event) {
|
|
|
<div class="overlay"></div>
|
|
|
<script src="assets/js/main.js"></script>
|
|
|
</body>
|
|
|
-</html>
|
|
|
+</html>
|